Uncovering metadata from semi-structured cohort data
Harmonisation of attributes across different cohorts is very challenging and labour intensive, but critical to leverage the collective potential of the data. While this problem is common across different domains, their individual needs and requirements make it difficult to employ a common set of tools or to directly apply standard machine learning techniques. For example, besides well structured metadata, cohorts are also described in scientific publications, and supplementary sections of research papers often contain large amounts of semi-structured data. This supplementary data could contain additional information which could both reaffirm and enhance original data and thus would be very helpful in search and data discovery. Due to its nature and scale, this data is not amenable to manual processing, and instead requires application of machine learning and Natural Language Processing (NLP) techniques to extract any useful information.
The CINECA text mining group aims to provide common tools and methods to extract additional metadata from structured and semi-structured fields in cohorts’ data.
Text mining team
The text-mining team encompasses members from CINECA participant institutions such as SFU, EMBL-EBI, HES-SO, and UCT. The teams are initially focusing on building tools based on their prior experience/knowledge targeting specific cohorts. The goal is to expose each of them as (micro)services, which will then be made available via a collective interface for general cohort text analysis related tasks.
Focusing on the CoLaus/PsyCoLaus cohort data, HES-SO/SIB has developed a pipeline using MetaMap, a machine learning and rule based framework for assigning unambiguous metadata descriptors to free text. SFU has developed a rule-based text mining tool LexMapr that cleans up and parses shorter form unstructured text to extract biomedical entities and map these to standard ontology terms. EMBL-EBI has developed Zooma and Curami pipelines to annotate and curate semi-structured data, which we describe further below.
Zooma pipeline
Zooma is an ontology annotation tool developed at EMBL-EBI for mapping free text to ontology terms. Zooma is backed by a linked data repository of annotation knowledge which contains curated annotations derived from many publicly available data sources such as Expression Atlas, Open Targets and GWAS Catalog. Therefore Zooma can facilitate annotations relating to a diverse range of topics including disease and phenotypes, drug treatments, anatomical components, species, cell types, etc. Furthermore Zooma can be easily configured to use new data sources or prioritise certain data sources over others to enhance context sensitivity. To increase FAIRness (1) of its data, BioSamples has developed a pipeline to automatically annotate sample attribute-values with ontologies using Zooma. This allows researchers to do complex queries using ontology expansion and synonyms - for example, searching for heart diseases will return samples annotated with myocardial infarction using ontology expansion.
Figure 1: Ontology annotation of Sample attribute-value using Zooma
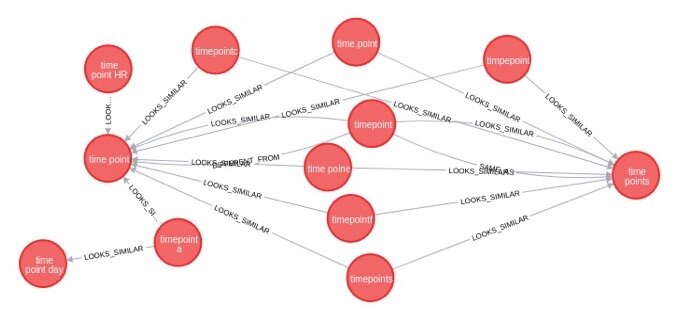
Curami pipeline
Curami is a machine assisted curation tool for harmonising data in the EBI BioSamples database. Curami aims to both identify erroneous attributes in the dataset and find a suitable replacement for it. Attribute pairing strategy is based on the assumption that, in most cases, the replacement for an erroneous attribute can be found within the dataset. After the text normalisation process Curami applies different NLP techniques to identify semantically and syntactically similar pairs of attributes. Filtered attribute pairs are loaded into neo4j graph database for identifying clusters of similar attributes. Sorted pairs based on curation impact are manually curated before feeding back to the BioSamples database. In the future we expect to use these curation results for machine learning analysis.
Figure 2: Neo4j cluster for lexically similar pairs
Conclusion
Using Zooma we have been able to add 15 million ontology annotations onto the 14 million public samples that BioSamples contains as of September 2020. We will apply Zooma for annotating cohort data with existing and new ontologies developed as a part of CINECA project to streamline mapping to the CINECA metadata model and improve data integration. Curami has allowed for automated curation of in excess of 40 million attributes. We recently deployed a curami-based recommendation service to improve the comprehensiveness of the metadata at submission time. Utilised together, Zooma and Curami increase data FAIRness at scale both on existing data and for future submissions.